RCSB PDB Data API
The RCSB PDB offers two ways to access data through application programming interfaces (APIs):
- REST-based API: refer to the full REST-API documentation
- GraphQL-based API: use in-browser GraphiQL tool to refer to the full schema documentation
Stay current with API announcements by subscribing to the RCSB PDB API mailing list:
- signing in with existing google account and subscribe
- or send an email to api+subscribe@rcsb.org
REST-based API
The REST-based API supports the HTTP GET method to access the PDB data through a set of endpoints (or URLs). See Data Organization section for more information on the underlying data organization.
GET request
The path of the endpoints starts with https://data.rcsb.org/rest/v1/core, followed by the type
of the resource, e.g. entry, polymer_entity, and the identifier. Note, that compound
identifiers, such as entity ID, assembly ID, and entity instance ID (or chain ID) are passed as path parameters.
Example request endpoints:
- https://data.rcsb.org/rest/v1/core/entry/4HHB - here "4HHB" is an alphanumeric identifier of PDB structure (PDB ID).
- https://data.rcsb.org/rest/v1/core/polymer_entity/4HHB/1 - here "4HHB" is a PDB ID and 1 is a unique identifier of an entity in PDB structures.
- https://data.rcsb.org/rest/v1/core/polymer_entity_instance/4HHB/A - here "4HHB" is a PDB ID and "A" is a unique chain ID (one or multiple alphanumeric characters; e.g., A, AA). Note, that here chain ID corresponds to _label_asym_id in PDBx/mmCIF schema.
Response
For any given request, if the data is found on the server, the API will return HTTP response code
200 (OK) – along with the response body in JSON format. For more information on the respond
schema see the REST-API documentation or refer to the
Data Schema section of this tutorial.
In case data is NOT found on the server (e.g.
https://data.rcsb.org/rest/v1/core/entry/xxxx) or the requested endpoint could not be found
(e.g.
https://data.rcsb.org/rest/v1/core/foo), then the API will return HTTP response code 404
(Fot Found).
GraphQL-based API
GraphQL server operates on a single URL/endpoint, https://data.rcsb.org/graphql, and all
GraphQL requests for this service should be directed at this endpoint. GraphQL HTTP server handles the
HTTP GET and POST methods.
GET request
If the "query" is passed in the URL as a query parameter, the request will be parsed and handled as the HTTP GET request. For example, to execute the following GraphQL query:
This query string should be sent via an HTTP like so:
In the example above, the query arguments are written inside the query string. The query
arguments can also be passed as dynamic values that are called variables. The variable
definition looks like ($id: String!) in the example below. It lists a variable, prefixed by $,
followed by its type, in this case String (! indicates that a non-null argument is required).
The following is equivalent to the previous query:
With variable defined like so:
Query variables, in this case, should be sent as a URL-encoded string in an additional query parameter
called variables.
POST request
The GraphQL server accepts POST requests with a JSON-encoded body. A valid GraphQL POST request should use
the application/json content type, must include query, and may include variables.
Here's an example for a valid body of a POST request:
Response
Regardless of the method by which the query and variables were sent, the response is returned in JSON format. A query might result in some data and some errors. The successful response will be returned in the form of:
Error Handling
Error handling in REST is pretty straightforward, we simply check the HTTP headers to get the status of a
response. Depending on the HTTP status code we get ( 200 or 404), we can easily
tell what the error is and how to go about resolving it. GraphQL server, on the other hand, will always
respond with a 200 OK status code. When an error occurs while processing GraphQL queries, the
complete error message is sent to the client with the response. Below is a sample of a typical GraphQL error
message when requesting a field that is not defined in the GraphQL schema:
Using GraphQL vs REST API
REST API offers a simple and easy-to-use way to fetch the data and returns a fixed data structure. If you need a full set of fields for a given object in the macromolecular data hierarchy, the REST API may be a better fit. GraphQL enables declarative data fetching and gives power to request exactly the data that is needed. Also, GraphQL query allows you to traverse the entire hierarchy of the macromolecular data in a single request. Conversely, with the REST API multiple round trips are needed to fetch the data from different levels in the macromolecular hierarchy.
No matter which method is used, the data returned by the REST API and the GraphQL query will be identical as they query the same source.
Data Organization
Biological molecules have a natural structural hierarchy, building from atoms to residues to chains to assemblies. The following definitions are relevant to the way the atomic coordinates, experimental data, and metadata are organized for each structure:
| Level | Description |
|---|---|
Entry |
Annotations pertaining to a particular structure (entry), designated with an alphanumeric entry ID (PDB ID, e.g. 1Q2W or CSM ID, e.g. AF_AFP68871F1). Annotations include the title of the entry, list of depositors, date of deposition, date of release, experimental details, etc. |
Entity |
Annotations describe the distinct (chemically unique) molecules present in entries. Three
types of entities are differentiated:
|
Entity Instance |
Entity instances (also referred to as "chains") are distinct copies of entities present in entries.
There can be multiple instances of a given entity. Entity instance data contains information that
can differ for each instance. For example, structural connectivity, secondary structure, validation
data, etc. Note, that information common for all copies of the same molecule is stored at the entity
level. Similarly to entity data, three types of entity instances are differentiated:
polymer_entity_instance, branched_entity_instance,
nonpolymer_entity_instance.
|
Assembly |
Annotations describe structural elements that form a biological assembly (also sometimes referred to as the biological unit), such as transformations required to generate the biological assembly, the information regarding the evidence of assembly, the annotations on the symmetry of polymeric subunits, etc. |
Chemical Component |
Chemical components describe all residues and small molecules found in entries. The annotations at this level include chemical descriptors (SMILES & InChI), chemical formula, systematic chemical names, etc. |
Data Schema
All data stored in the PDB archive conform to the PDBx/mmCIF data dictionary. This data is augmented with annotations coming from external resources and computed data. The RCSB PDB data representation, powered by the JSON Schema language, is connected to the data hierarchy. Such data organisation groups annotations in objects defined as follows:
- Entry Schema
- Polymer Entity Schema
- Branched Entity Schema
- Non-polymer Entity Schema
- Polymer Instance Schema
- Branched Instance Schema
- Non-polymer Instance Schema
- Assembly Schema
- Chemical Component Schema
Typically, integrated data will be added as additional fields to any of the objects above. Some data, however, has a substantial overlap with the source data in terms of content. Such data appears as a separate object with dedicated schema, where original semantics preserved as much as possible:
The relationships between these objects are explicitly implemented through attributes in a dedicated
container object: rcsb_[...]_container_identifiers, where [...] should be replaces
with the type of the object, e.g. entry, polymer_entity, assembly.
For example, rcsb_entry_container_identifiers contains polymer_entity_ids, branched_entity_ids, non_polymer_entity_ids attributes that hold corresponding entity IDs.
GraphQL Schema
All GraphQL queries are validated and executed against the GraphQL schema. The GraphQL schema contains nodes and edges, where nodes being objects, that represent macromolecular data hierarchy, and edges being the relationships between those objects. See Nodes and Edges for more details.
You can use GraphiQL, which is a "graphical interactive in-browser GraphQL IDE", to explore GraphQL schema. It lets you try different queries, helps with autocompletion and built-in validation. The collapsible Docs panel (Documentation Explorer) on the right side of the page allows you to navigate through the schema definitions. Click on the root Query link to start exploring the GraphQL schema.
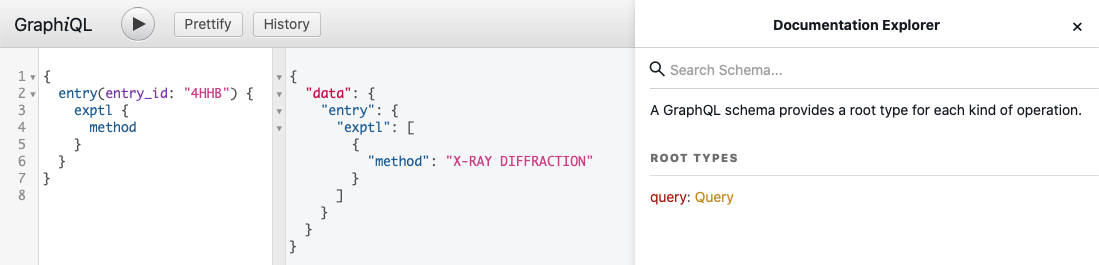
Fetch GraphQL Data
You can use GraphQL to fetch data for objects from different levels of data organisation with a single API call. GraphQL is strongly typed. It means queries are executed within the context of a data schema and only queries to valid fields will be successfully processed.
Root Queries
Root queries define entry-points from where you can start traversing the data hierarchy. You can start your query from any object in the hierarchy and visit adjacent objects through bidirectional links (edges) connecting nodes. See Nodes and Edges for more details.
Root queries have parameters and except either a single identifier for requested object (e.g. entry ID, entity ID, etc.) or multiple identifiers supplied as a list. The following example shows how to fetch experimental method name for multiple PDB entries:
When requesting data for multiple objects compound identifiers should follow the format:
- [entry_id]_[entity_id] - for polymer, branched, or non-polymer entities (e.g. 4HHB_1)
- [entry_id].[asym_id] - for polymer, branched, or non-polymer entity instances (e.g. 4HHB.A)
- [entry_id]-[assembly_id] - for biological assemblies (e.g. 4HHB-1)
For example:
Nodes and Edges
One of the benefits of GraphQL is that it simplifies traversing the graph of relationships between
different objects. In RCSB Data API relationships are modelled with data Node objects connected
through Edges links. For example, CoreEntry and CorePolymerEntity are data nodes that
are connected through polymer_entities link that allows fetching the data all polymer entities
present in a given entry.
Node is an object that holds all fields for a given level in the data hierarchy. Nodes have fields that can be complex objects or scalar values. GraphQL queries are built by specifying fields within fields (also called nested subfields) until only scalars are returned.
Edges represent connections between nodes. Through edges the API allows you to traverse the data
hierarchy by visiting adjacent data objects, e.g. from entry to polymer_entity,
from polymer_entity to polymer_entity_instance, etc. Traversing up the hierarchy
is also possible. For example, you can fetch an organism name for a given polymer entity using the
polymer_entity root query and in the same query fetch an experimental method name, that
resides at the entry level, using the entry edge.
Usage Guidelines
There are currently no limits set on the requests rate or query complexity. However, complex queries for a large number of objects are bound to have performance issues. To prevent the API from being overwhelmed and improved the performance of your queries check out the suggestions below.
Batch Large Requests
Requesting a large number of objects at a time is deemed resource intensive and not recommended. Making requests in periodic batches, instead of a single request for a large number of objects, can be more effective.
GraphQL endpoints require to explicitly specify ID(s) for the requested data objects, there are no endpoints to request all data objects. The Repository Holdings Service REST API current entries endpoint provides a full list of current PDB IDs.
Cache Data For Repeat Calls
Repeat calls to PDB data within a weekly update window should be cached when possible.
Data Attributes
The RCSB PDB data available through the APIs includes only commonly used annotations, rather than supporting all metadata available in the PDBx/mmCIF data dictionary. Refer to the Data Attributes page for a full list of objects and their attributes.
API Clients
Python
The rcsb-api package
provides a Python interface to the RCSB PDB Search and Data APIs (an overview has been published in
Journal for Molecular Biology).
Use the rcsbapi.search module to fetch lists of PDB IDs corresponding to advanced query searches,
and the rcsbapi.data module to fetch data about a given set of structure IDs. RCSB PDB maintains
the current version of this package on GitHub.
You can find example use cases demonstrating how to utilize this package in scripting workflows in the py-rcsb-api GitHub repository. These examples provide practical implementations of common tasks, helping users understand how to integrate the package into their own applications. The notebooks serve as a reference for building custom workflows using RCSB resources.
Examples
This section contains additional examples for using the GraphQL-based RCSB PDB Data API.
Entries
Fetch information about structure title and experimental method for PDB entries:
Primary Citation
Fetch primary citation information (structure authors, PubMed ID, DOI) and release date for PDB entries:
Polymer Entities
Fetch taxonomy information and information about membership in the sequence clusters for polymer entities:
Polymer Instances
Fetch information about the domain assignments for polymer entity instances:
Note, that label_asym_id is used to identify polymer entity instances.
Carbohydrates
Query branched entities (sugars or oligosaccharides) for commonly used linear descriptors:
Sequence Positional Features
Sequence positional features describe regions or sites of interest in the PDB sequences, such as binding sites, active sites, linear motifs, local secondary structure, structural and functional domains, etc. Positional annotations include depositor-provided information available in the PDB archive as well as annotations integrated from external resources (e.g. UniProtKB).
Positional features are available for polymer_entities or polymer_entity_instances
data objects (see Data Organization section for more information on the data organization).
Polymer entity annotations are obtained from sequence alone (e.g. modified monomers) and polymer entity
instance annotations from 3D structural information (e.g., the secondary structure content of proteins).
This example queries polymer_entity_instances positional features. The query returns features
of different type: for example, CATH and SCOP classifications assignments integrated from UniProtKB data,
or the secondary structure annotations from the PDB archive data calculated by the data-processing program
called MAXIT (Macromolecular Exchange and Input Tool) that is based on an earlier ProMotif implementation.
Reference Sequence Identifiers
This example shows how to access identifiers related to entries (cross-references) and found in data collections other than PDB. Each cross-reference is described by the database name and the database accession. A single entry can have cross-references to several databases, e.g. UniProt and GenBank in 7NHM, or no cross-references, e.g. 5L2G:
Chemical Components
Query for specific items in the chemical component dictionary based on a given list of CCD ids:
Computed Structure Models
This example shows how to get a list of global Model Quality Assessment metrics for AlphaFold structure of Hemoglobin subunit beta:
Integrative Structures
Dara API delivers integrative structures alongside the experimental structures and computed (predicted) models. Integrative structures are determined using a combination of data from traditional techniques such as X-ray crystallography, NMR spectroscopy, and 3D Electron Microscopy (3DEM) along with other experimental approaches like Small Angle Scattering (SAS), Crosslinking Mass Spectrometry, Atomic Force Microscopy, Forster Resonance Energy Transfer, and more. These methods help build structure models of large, dynamic, and heterogeneous biological assemblies that are difficult to resolve with a single technique.
This example shows how to query integrative structures to get information on model composition, input datasets used in integrative modeling, and source databases for the input datasets:
Archive-wide Queries
Performing queries across all currently released PDB structures involves multiple steps, as the process is designed to efficiently handle large-scale data. Follow the steps below:
- Retrieve the complete list of current PDB IDs. This can be done using the Holdings REST API. The API will return a JSON array containing all current PDB IDs
- To efficiently query the dataset and avoid overwhelming the GraphQL endpoint, batch the PDB IDs into smaller chunks
- Once you have your batches of PDB IDs, use them to query the GraphQL endpoint
Explore this GitHub repository for Python notebooks and scripts demonstrating real use cases of this multi-step querying process.
Migrating from Legacy Fetch API
Applications written on top of the Legacy Fetch APIs no longer work because these services have been discontinued. This migration guide describes the necessary steps to convert applications from using Legacy Fetch API Web Service to a new RCSB Data API.
Acknowledgements
To cite this service, please reference:
- Yana Rose, Jose M. Duarte, Robert Lowe, Joan Segura, Chunxiao Bi, Charmi Bhikadiya, Li Chen, Alexander S. Rose, Sebastian Bittrich, Stephen K. Burley, John D. Westbrook. RCSB Protein Data Bank: Architectural Advances Towards Integrated Searching and Efficient Access to Macromolecular Structure Data from the PDB Archive, Journal of Molecular Biology, 2020. DOI: 10.1016/j.jmb.2020.11.003
Related publications:
- H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne. (2000) The Protein Data Bank Nucleic Acids Research, 28: 235-242.
- Stephen K Burley, Helen M. Berman, et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy (2019) Nucleic Acids Research 47: D464–D474. doi: 10.1093/nar/gky1004.
Contact Us
Contact info@rcsb.org with questions or feedback about this service.